- Head of research group
HIGH THROUGHPUT SEQUENCING – METAGENOMICS- VIRAL GENOMICS
1. Use of high throughput techniques (next generation sequencing) in the detection of known and novel viral pathogens in acute and chronic diseases
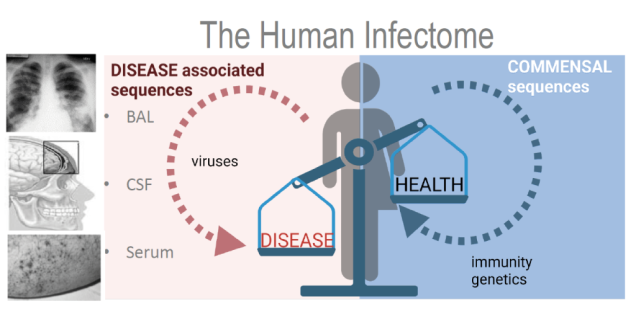
Numerous infectious diseases emerged or re-emerged over the past decades. Pathogen identification/detection methods have significantly improved over time; however sample preparation (nucleic acid extraction), development of reliable diagnostic tools as well as proving causality (Hill’s criteria) once a pathogen has been identified are still challenging. Next generation sequencing together with PCR amplification techniques are used to hunt for new viruses in acute and chronic diseases with putative infectious etiology.
Improvements of sample preparation for successful detection of viral pathogens from diagnostic samples, development of reliable protocols suitable for detection of pathogens from clinical samples using next generation sequencing together with Capture sequencing approaches are key elements in our research.
Together with the Research group for Virus Genomics, Leibniz Institute for Experimental Virology (HPI), Hamburg we develop standardized NGS protocols and procedures for the unbiased detection of viruses, bacteria, fungi and parasites in diagnostic samples. Together with our collaborators at the HPI we optimize and adapt bioinformatic methods to detect and analyze pathogens in clinical samples of known or suspected infectious etiology. All protocols and methods are developed in close cooperation between clinical and research groups at the partner institutions.
Selected publications:
-
Lopez-Labrador FX, Brown JR, Fischer N, et al.
Recommendations for the introduction of metagenomic high-throughput sequencing in clinical virology, part I: Wet lab procedure. J Clin Virol. 2021;134:104691.
-
de Vries JJC, Brown JR, Couto N, et al.
Recommendations for the introduction of metagenomic next-generation sequencing in clinical virology, part II: bioinformatic analysis and reporting. J Clin Virol. 2021;138:104812.
-
de Vries JJC, Brown JR, Fischer N, et al.
Benchmark of thirteen bioinformatic pipelines for metagenomic virus diagnostics using datasets from clinical samples. J Clin Virol. 2021;141:104908.
-
Alawi M, Burkhardt L, Indenbirken D, et al.
DAMIAN: an open source bioinformatics tool for fast, systematic and cohort based analysis of microorganisms in diagnostic samples. Sci Rep. 2019;9(1):16841.
-
Gunther T, Haas L, Alawi M, et al.
Recovery of the first full-length genome sequence of a parapoxvirus directly from a clinical sample. Sci Rep. 2017;7(1):3734.Baechlein C, Fischer N, Grundhoff A, et al. Identification of a Novel Hepacivirus in Domestic Cattle from Germany. J Virol. 2015;89(14):7007-7015.
-
Fischer N, Indenbirken D, Meyer T, et al.
Evaluation of Unbiased Next-Generation Sequencing of RNA (RNA-seq) as a Diagnostic Method in Influenza Virus-Positive Respiratory Samples. J Clin Microbiol. 2015;53(7):2238-2250.
-
Fischer N, Rohde H, Indenbirken D, et al.
Rapid metagenomic diagnostics for suspected outbreak of severe pneumonia. Emerg Infect Dis. 2014;20(6):1072-1075.
2. SARS CoV-2 genomics: Monitoring the Pandemic in the metropolitan region of Hamburg
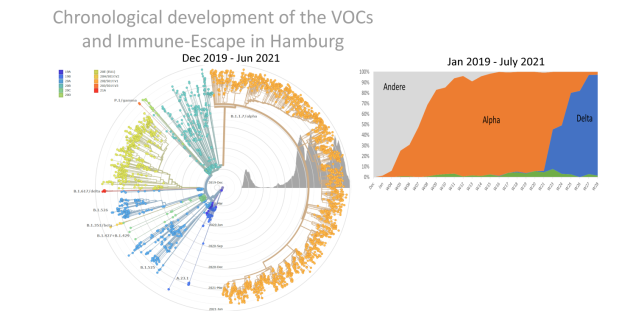
The recent emergence of SARS-CoV-2 variants with potentially increased transmissibility (i.e., the alpha variant of the B.1.1.7 lineage, the beta variant of the B.1.351 lineage, the gamma variant of the P.1 lineage and recently the current pandemic dominating variant, the delta variant of the B.1.617.2 lineage) clearly emphasize the importance of tracking the emergence of variants and their spread.
With the support of the Hanseatic City of Hamburg, we sequence randomly selected SARS-CoV-2 samples, dependent on the number of positive cases occurring between 5 - 30%. The sequence data are analyzed using computer-assisted methods and subsequently evaluated by a joint HPI/UKE team of experts in order to be able to detect at an early stage the spread of already known mutations, but also the emergence of possible new ones. Since the beginning of the Corona pandemic, we have sequenced more than 5,000 SARS-CoV-2 genomes. Sequence data have been collected from a systematic cross-section of all SARS-CoV-2 samples received by UKE to monitor virus arrivals in Hamburg, track the spread of SARS-CoV-2, and, in particular, to closely monitor and evaluate genetic changes of the pathogen in the metropolitan region. Sequence analyses commissioned by Hamburg health departments and authorities have also already been able to make an important contribution in the investigation of local outbreaks in hospitals, schools, daycare centers and nursing homes.
The Hamburg viral genomics surveillance platform is also an important component of the collection of SARS-CoV-2 sequence data nationwide. The data will also be forwarded to the Robert Koch Institute (RKI) in accordance with the federal CorSuRV regulation, which has been in effect since January 18, 2021.
The most recent data on this can be found at the following link .
Monitoring mutations in longitudinal samples of patients with prolonged disease courses
The fact that the first described variant of concern (VOC), the alpha variant appears to have acquired an unusual high number of mutations within a short time period has led to the speculation that this variant may have rapidly evolved during chronic infection of an immunocompromised patient. This hypothesis is largely based on few reports on only a small number of patients suffering from prolonged infection. However, the dynamics with which novel variants evolve or emerge during the millions of ‘conventional’ infection and transmission events occurring worldwide are largely unknown. It is of high interest to i) determine frequency, type and potential origin of variants that emerge during transmission in the general population, and ii) to investigate whether immunocompromised patients with prolonged infections indeed display fundamentally increased intra-host evolution that may allow more rapid emergence of SARS-CoV-2 variants. Together with our collaborators at the HPI and collaborators in the clinics we investigate intra-host genetic diversity in longitudinal samples from immune-compromised patients (+/- antiviral treatment) to evaluate emergence of mutations with and without selection pressure.
Selected publications:
- Brehm, T. T. et al. SARS-CoV-2 Reinfection in a Healthcare Worker Despite the Presence of Detectable Neutralizing Antibodies. Viruses 13, doi:10.3390/v13040661 (2021).
- Heinrich, F. et al. Dying of VOC-202012/01 - multimodal investigations in a death case of the SARS-CoV-2 variant. Int J Legal Med, doi:10.1007/s00414-021-02618-8 (2021).
- Nörz, D. et al. Evaluation of a fully automated high-throughput SARS-CoV-2 multiplex qPCR assay with built-in screening functionality for del-HV69/70- and N501Y variants such as B.1.1.7. J Clin Virol 141, 104894, doi:10.1016/j.jcv.2021.104894 (2021).
- Pfefferle, S. et al . SARS Coronavirus-2 variant tracing within the first Coronavirus Disease 19 clusters in northern Germany. Clin Microbiol Infect 27, 130 e135-130 e138, doi:10.1016/j.cmi.2020.09.034 (2021).
- Günther, T. et al. SARS-CoV-2 outbreak investigation in a German meat processing plant. EMBO Mol Med 12, e13296, doi:10.15252/emmm.202013296 (2020).
HUMAN POLYOMAVIRUSES
CHRONIC/PERSISTENT INFECTIONS - PATHOGENESIS
1. Viral Tumorigenesis: How Merkel cell polyomavirus contributes to Merkel cell cancer tumorigenesis und tumor progression
Merkel cell polyomavirus (MCPyV) is one out of seven known human tumor viruses. The virus was initially identified in 2009 in Merkel cell cancer (MCC) tissue samples by Juan Chang and Patrick Moore, who also identified the Kaposi-sarcoma-associated virus (KSHV). Multiple groups confirmed that most MCCs are MCPyV positive and that the virus is causally contributing to MCC tumorigenesis. Tumor cells display a monoclonal integration of the viral DNA in the host genome. Tumor cell viability is dependent on viral oncoproteins large T (LT) and small T-antigen (sT), and integrated viral genomes carry tumor-specific mutations not present in the viral episomes rescued from healthy people. Low mutational burden and relatively stable genomes in virus + MCCs with hardly any typical cancer-driving alterations indicate that the viral oncoproteins LT/sT mainly drive tumorigenesis. The dependence of tumor cells on viral oncoprotein expression and that only a few mutations are present in the host genome make the viral oncoproteins and their function ideal new targets to find new therapeutic options against MCC:
Our research focuses on how the viral oncoproteins sT and LT contribute to tumorigenesis and to MCC progression. We focus on the interaction of viral oncoproteins and the tumor microenvironment as well, as we are interested in the viral-induced changes on host chromatin and gene expression.
Selected publications:
- Czech-Sioli, M. et al . High-resolution analysis of Merkel Cell Polyomavirus in Merkel Cell Carcinoma reveals distinct integration patterns and suggests NHEJ and MMBIR as underlying mechanisms. PLoS Pathog 16, e1008562, doi:10.1371/journal.ppat.1008562 (2020).
- Riethdorf, S. et al. Detection and Characterization of Circulating Tumor Cells in Patients with Merkel Cell Carcinoma. Clin Chem 65, 462-472, doi:10.1373/clinchem.2018.297028 (2019)
- Knips, J. et al. Spontaneous lung metastasis formation of human Merkel cell carcinoma cell lines transplanted into scid mice. Int J Cancer 141, 160-171, doi:10.1002/ijc.30723 (2017).
- Grundhoff, A. & Fischer, N. Merkel cell polyomavirus, a highly prevalent virus with tumorigenic potential. Curr Opin Virol 14, 129-137, doi:10.1016/j.coviro.2015.08.010 (2015).
- Borchert, S. et al. High-affinity Rb binding, p53 inhibition, subcellular localization, and transformation by wild-type or tumor-derived shortened Merkel cell polyomavirus large T antigens. J Virol 88, 3144-3160, doi:10.1128/JVI.02916-13 (2014).
2. Polyomavirus persistence
Chronic viral infections are a major challenge in our increasingly aging society. Many of these infections can be reactivated in an immunocompromised host and result in severe disease. In particular, herpesviruses (e.g., HSV, CMV, and EBV) are known to infect humans and subsequently remain persistent. Under immunosuppression, they can then contribute to severe, life-threatening diseases, such as inflammation of the central nervous system or, in the case of EBV and KSHV, tumorigenesis. There is no therapeutic option to eradicate a latent infection. Similar to herpesviruses, other nuclear DNA viruses establish a lifelong infection, a chronic infection: Papillomaviruses and Polyomaviruses. These viruses also contribute to severe diseases, including malignancies. Polyomaviruses, which are clinically relevant, are BK virus, JC Virus, and Merkel cell polyomavirus.
While herpesviruses establish a particular form of chronic infection, viral latency, in which hardly any viral proteins or viral RNAs are expressed, the chronic infection (called persistence) of polyomaviruses and papillomaviruses can take different forms, such as continuous re-infection of cells or maintenance of episomal viral genomes beyond cell division in the case of papillomaviruses.
Regardless of which specific form of viral persistence is established, this process requires selective suppression/tuning of viral gene expression, which we aim to answer for polyomaviruses.
We are pursuing these goals in the recently established
DFG research group FOR5200
DEEP-DV
:
Disrupt- Evade - Exploit: Gene expression and host response programming in DNA virus infection.
Sprecherinnen: Melanie Brinkmann/TU Braunschweig and Nicole Fischer/UKE
Selected publications:
- Czech-Sioli, M. et al. The Ubiquitin-Specific Protease Usp7, a Novel Merkel Cell Polyomavirus Large T-Antigen Interaction Partner, Modulates Viral DNA Replication. J Virol 94, doi:10.1128/JVI.01638-19 (2020).
- Siebels, S. et al. Merkel Cell Polyomavirus DNA Replication Induces Senescence in Human Dermal Fibroblasts in a Kap1/Trim28-Dependent Manner. mBio 11, doi:10.1128/mBio.00142-20 (2020). Fibroblasts in a Kap1/Trim28-Dependent Manner. mBio 11, doi:10.1128/mBio.00142-20 (2020).
- Theiss, J. M. et al. A Comprehensive Analysis of Replicating Merkel Cell Polyomavirus Genomes Delineates the Viral Transcription Program and Suggests a Role for mcv-miR-M1 in Episomal Persistence. PLoS Pathog 11, e1004974, doi:10.1371/journal.ppat.1004974 (2015).
3. Antivirals against human polyomaviruses, e.g. BKV
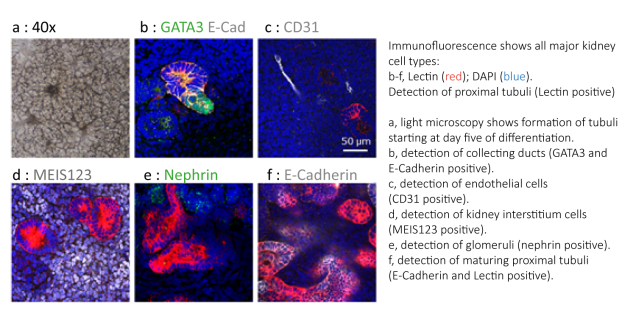
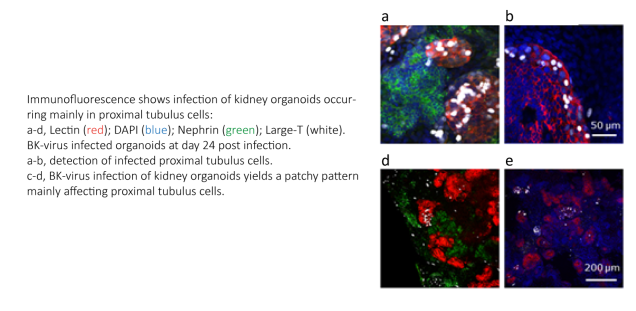
Human polyomaviruses (HPyV) are highly abundant, persist for life and can induce severe and life threatening disease in elderly and immunosuppressed patients. Of the 14 known human polyomaviruses, four are of clinical relevance: BK Virus (BKV), JC Virus (JCV), Trichodysplasia spinulosa-associated polyomavirus (TSPyV) and Merkel cell polyomavirus (MCPyV). In particular, BKV and JCV re-activation in immunosuppressed patients can be life threatening: BKV reactivation is a major complication in immunosuppressed patients, with uncontrolled BKV replication resulting in haemorrhagic cystitis in 5 – 15% of allogenic transplant patients, and up to 10% of kidney transplant patients suffering from polyomavirus-associated nephropathy (PVAN) and subsequent graft loss. As no specific viral inhibitors are available, treatment of PyV associated complications is presently limited to alleviation of the immunosuppressive regimen. To fill this gap we are performing high throughput sceens using small molecule inhibitor libraries to identify putative antivirals against SV40/BKV/JCV related viruses. We conduct detailed viral/genetic and structural studies, to address the mode of action of selected inhibitors and to establish a human kidney organoid system for close to in vivo testing of selected inhibitors.
This project is integrated in the German Center for Infection Research, DZIF, thematic translational unit Infection of the Immunocompromised host ( TTU IICH ).
Selected recent publications:
-
Koyro, T. F. et al.
Upregulation of HLA-F expression by BK polyomavirus infection induces immune recognition by KIR3DS1-positive natural killer cells. Kidney Int 99, 1140-1148, doi:10.1016/j.kint.2020.12.014 (2021).
Staff • Group Fischer

- Postdoc

- Doctoral student

- Postdoc

- Bioinformatics Facility
- Assistant physician
- Doctoral student